Arrival and departure
- Arrival, Sunday Nov. the 19th: a shuttle will run from the Ajaccio airport to the conference center. Estimated departure time: 5pm-6pm, depending on the flights of participants. Therefore, make sure to reach the Ajaccio before 5pm.
- Departure, Friday Nov. the 24th: a shuttle will bring back participants to the Ajaccio airport. Departure from the conference center circa 3pm. Therefore, take a flight after 5pm.
General program
- Morning 8h30-10h [break] 10:30 – 12:00 : Classroom lectures
- Afternoon : Practical work.
- Mo, We, Fr: 14:00 – 15:30 [ break] 16:00 – 17:30
- Tu, Th: 15:30-17:00 [break] 17:30-19:00
- Monday 17:45 – 19:00: Flash presentations by the participants
Program per day
Monday : The image alignment problem
Monday-bis : ≤ 5 min participants introduction
Tuesday : From raw data to context
Wednesday : Combining molecular dynamics and cryo-EM
Thursday : Flexibility in cryo-EM reconstruction
Friday : Hybrid methods for analyzing conformational variability in cryo-EM and cryo-ET data
… departure: Friday session ends early to allow bus to Ajaccio airport
Detailed program
Monday : The image alignment problem
Carlos Oscar Sozano – National Center of Biotechnology, Madrid, Spain
download slides- Context
- Goal functions
- Optimization strategies
- Deep learning and classical image processing
Monday, after the practical :
Participants Session. Short presentations (≤5 min) to tell about your research interests. Please have a couple of slides (pdf format) ready to project.Tuesday : From raw data to context
Christian Dienemann – MPI Gottingen, Germany
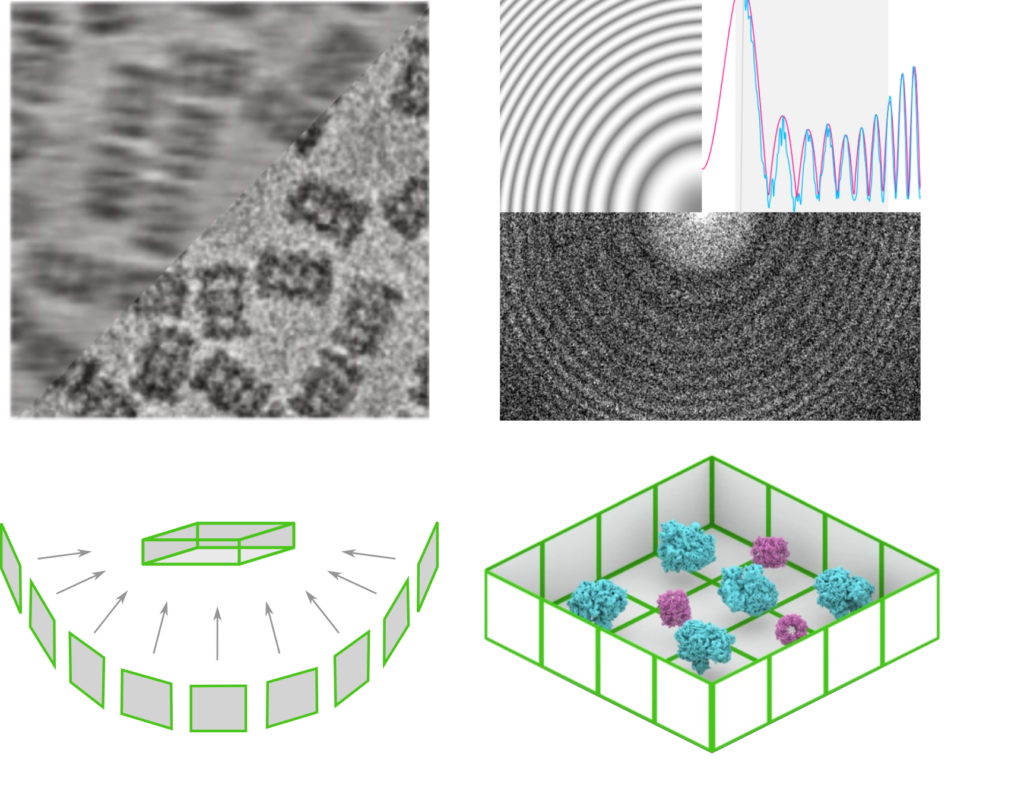
download slides- Preprocessing of cryo-EM images: methods for movie alignment, dose weighting and CTF-estimation
- Tomogram reconstruction: various reconstruction methods (SIRT, BP, IsoNet, etc.)
- Identification of image features: particle picking (2D and 3D) and segmentation
- Exploiting 3D context in tomograms: orientation priors, multi-species alignment
Wednesday: Combining molecular dynamics and cryo-EM
Pilar Cossio – Flatiron Institute, New-York, USA
download slides1 slides2- Basis of molecular dynamics
- Bayesian analysis
- Monte Carlo
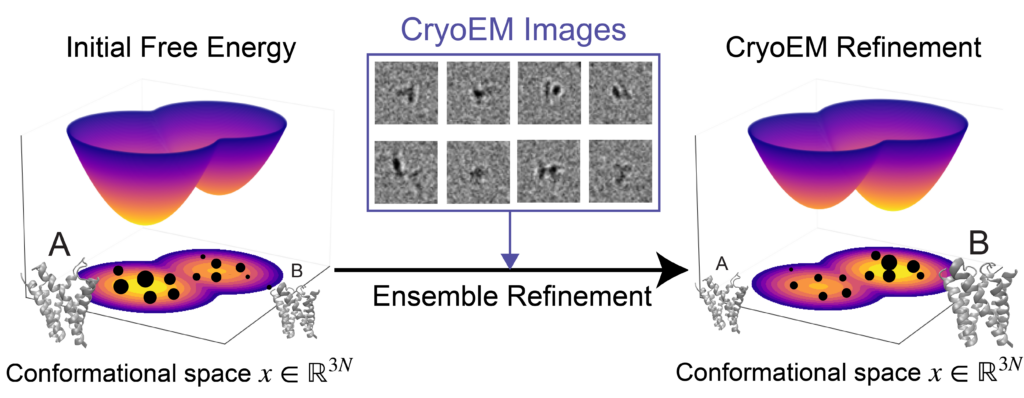
- Ensemble reweighting using cryoEM
Thursday: Flexibility in cryo-EM reconstruction
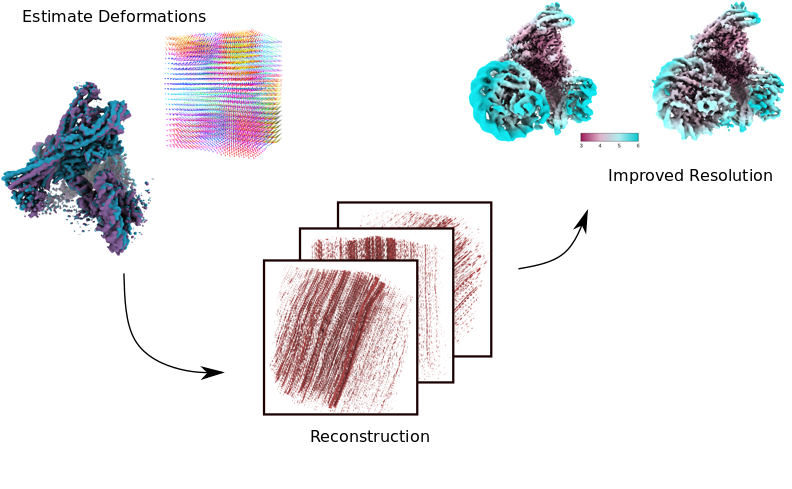
Johannes Schwab – Cambridge Univ, England
download slides- Mathematical formulation of 3D reconstruction in single particle cryo EM
- 3D reconstruction algorithms for homogeneous data
- Reconstruction methods for conformational heterogeneous data
- Estimating deformations for improved 3D reconstruction
Friday: Hybrid methods for analyzing conformational variability in cryo-EM and cryo-Et data
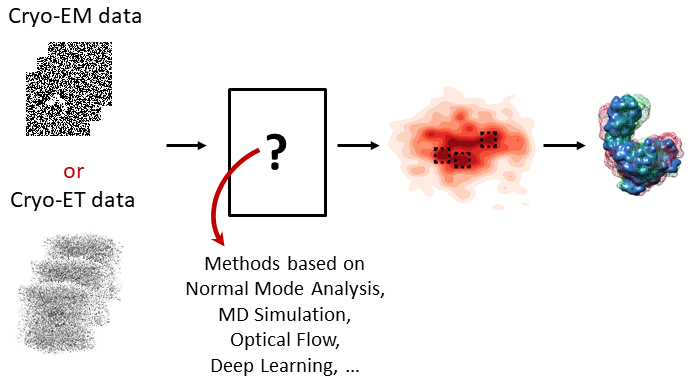
Slavica Jonic – CNRS, France
download slides and preprint- Cryo-EM vs. Cryo-ET: Principles, Advantages, Limitations
- Conformational changes of biomolecular complexes: Discrete vs. Continuous (Terminology, Examples)
- Conformational heterogeneity in cryo-EM and cryo-ET data: Obstacle and Opportunity
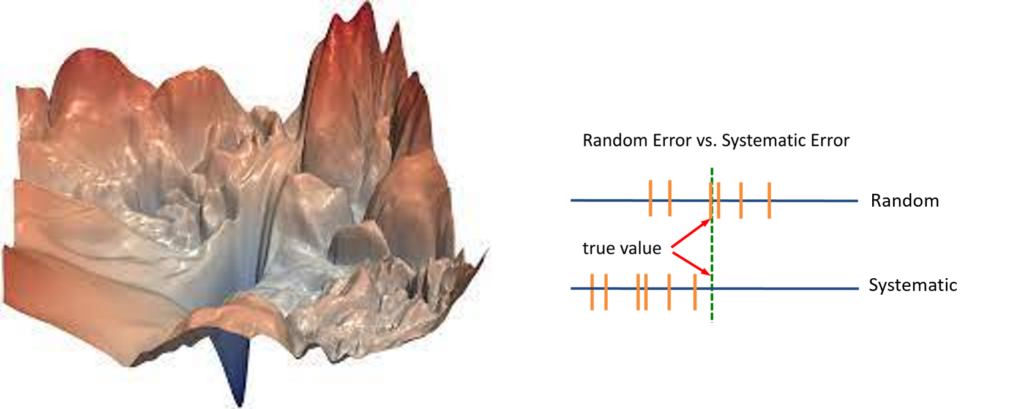
- Conformational landscapes and Free-Energy landscapes: Terminology, Examples
- Methods for analyzing conformational variability in cryo-EM and cryo-ET: Principles, Brief overview
- Combining dynamics simulation, image processing, and deep learning for analyzing conformational variability